Overview
The Mechanics & Devices Core provides state-of-the-art instrumentation, training, and resources to investigators studying the structure and functional properties of skeletal muscle during development and dysfunction with diseases. The Core can assess contractile function at the protein, filament, cell, tissue and organ levels, and provide assays for measuring the developing properties of early-stage muscle from animal and human sources such as inducible pluripotent stem cell derived muscle cells. The Core trains investigators individually and with workshops. We help investigators develop novel research assays and platforms for drug and small molecule testing and investigating engineered cells and tissues and gene therapies that are being developed for clinical application and commercialization.
Facilities and Resources Available

Tissue Mechanics: Measuring contractile properties in intact muscle and engineered muscle tissue.
We have multiple instruments that are capable of measuring tissue-level contractile mechanics and kinetics in intact rodent or engineered muscle tissues. Muscle tissues are hung between a length-controlling motor and a force transducer and are electrically stimulated to elicit contractions. These systems allow us to measure properties such as force-frequency relationships, force-length relationships, calcium handling, and others in live muscle tissue.
Single-Myocyte Mechanics: Intact or demembranated single-myocyte mechanics and kinetics
This system allows us to measure the contractile mechanics and kinetics from a single muscle cell, either membrane-intact or chemically demembranated. Similar to the tissue mechanics systems, individual myocytes are mounted between a length-controlling motor and a force transducer. Chemically demembranated preparations enable us to quantify changes in contraction force and kinetics in response to varying calcium concentrations, small-molecule drugs, or other compounds of interest. Membrane-intact cells are electrically stimulated, allowing us to measure similar properties as the tissue mechanics systems but at a single-cell level.

Myofibril Mechanics: Sub-cellular organelle (myofibril) contractile kinetics.
We reduce muscle cells down to their functional contractile organelle, the myofibril, which is composed of only a few sarcomeres in series. Myofibrils isolated from either human or animal myocytes are mounted between two glass microneedles, and contractile mechanics and kinetics are monitored during calcium-induced activation. Rapid length changes imposed on the myofibril reveal properties of crossbridge cycling kinetics, and rapid calcium removal provides information about myofilament relaxation.
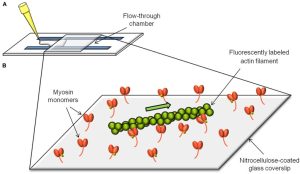
In vitro motility assay: contractile protein function
Fluorescently labeled thin filaments are visually monitored as they interact with an array of ATP-driven cycling myosin molecules, enabling quantification of filament sliding speed and efficiency. Thin filaments can either be bare F-actin or F-actin decorated with various regulatory proteins, which can include patient-specific regulatory protein variants. With this system, we can measure myosin function, thin filament regulation, and actomyosin interactions in healthy and diseased contexts.

Stopped-flow spectrometry: muscle protein chemo-mechanical analysis
This system measures the kinetics of protein-protein or protein-ligand interactions. We can quantify myosin crossbridge cycle kinetics, actin-myosin interaction rates, small molecule affinity for myofilament proteins, among many other important interactions in muscle biochemistry.
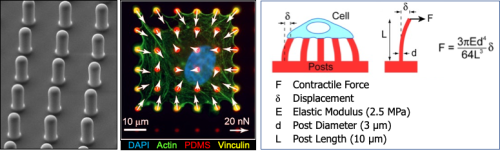
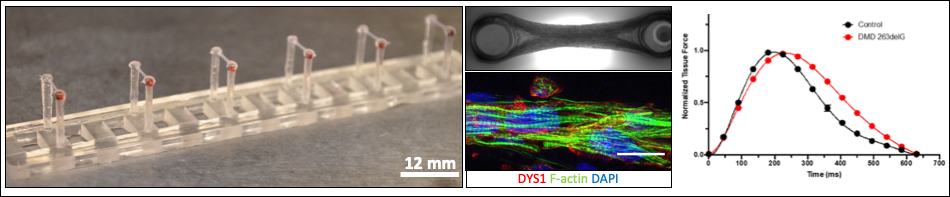
Muscle cell force assays using micropost arrays, black dot traction force microscopy, and engineered muscle tissue constructs.
Services
Interested in utilizing our services?
Please contact Core Director, Mike Regnier, and Program Manager, Katie Mitzelfelt (mregnier@uw.edu; kmitz@uw.edu), to discuss your project and learn more about how our services can support your project’s muscle mechanics and device needs.
Personnel
Dr. Michael Regnier is the Director of Core B. He is responsible for managing the scientific and financial operations of the Core. He directs the research activities of the Core and assists in the interpretation of the data from all work done by the Research Base and Center investigators. Dr. Regnier has over 30 years of experience analyzing contractile function in striated muscle and developing targeted therapeutic approaches to treating muscle disease.
Dr. Nathan Sniadecki is a Core B faculty lead who has developed approaches using micropost arrays, black dot traction force microscopy, and engineered muscle tissue constructs to assess the structural and functional properties of muscle cells. Dr. Sniadecki will supervise development of additional assays and protocols in the Core.
Dr. Joe Powers is a Core B faculty member who specializes in multiscale muscle biophysics and pathology. His lab develops experimental and computational approaches to investigate the biomechanics and mechanobiology of muscle cells in health and disease.
Dr. Tim McMillen is the lead Research Scientist for muscle mechanics in Regnier Lab. He has expertise in muscle mechanics experiments and Langendorff heart perfusions in conjunction with NMR to assess cardiac function, substrate utilization and energetics. He will train faculty, post-doctoral and graduate student investigators or work with them on Core B projects.
Ruby Padgett is a Research Scientist who works with Dr. Sniadecki on development and fabrication of muscle cell forces assays (micropost and/or ‘black dot’ traction force microscopy) and engineered muscle tissue force assays for measures of contractile capacity, muscle cell development and disease. She will work with collaborating investigators to develop novel tools and assays and be available to train others or perform experiments with them for Core B.
Overview
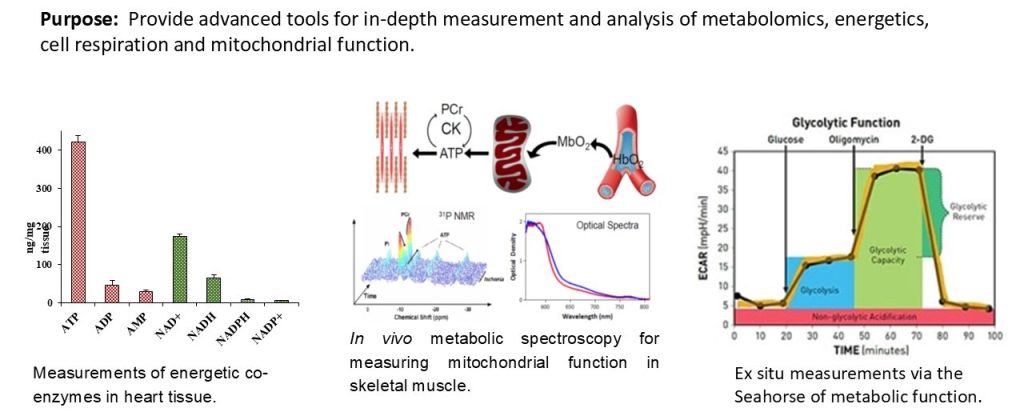
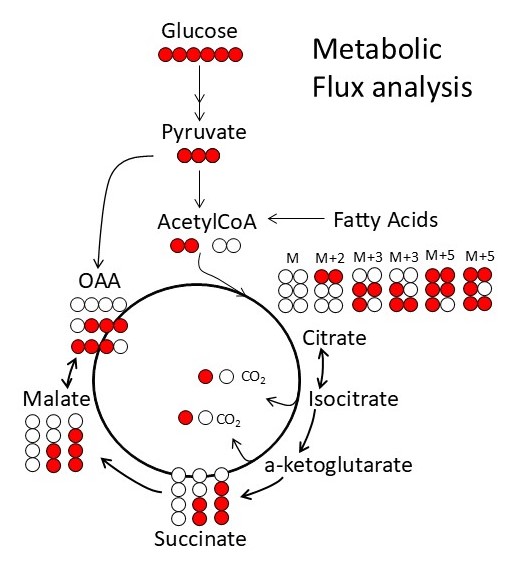
The Metabolism Core provides access to state-of-the-art instruments and unique expertise to characterize muscle metabolism and energetics. These tools include metabolomics analysis of muscle tissue, ligaments and tendons, in vivo measurements of mitochondrial function and energetics in mouse skeletal muscles, measurement of respiration, and mitochondrial function ex vivo in various muscle tissues. These tools will provide much greater insight into muscle biology, and an ability to monitor therapeutic approaches at the molecular level.
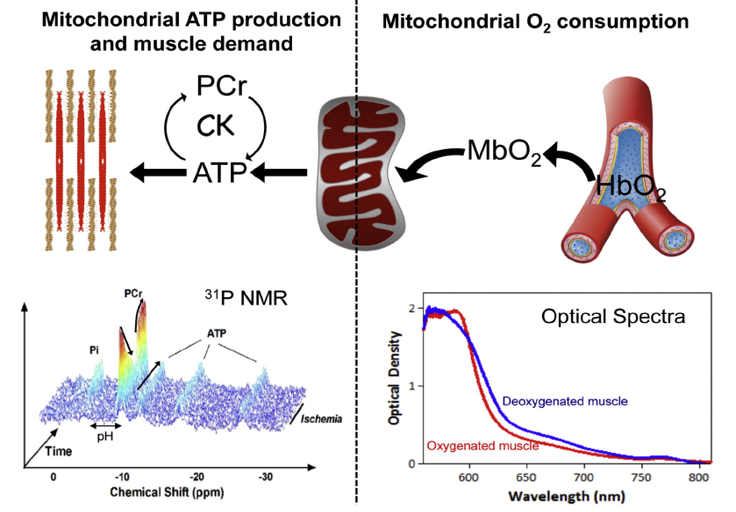
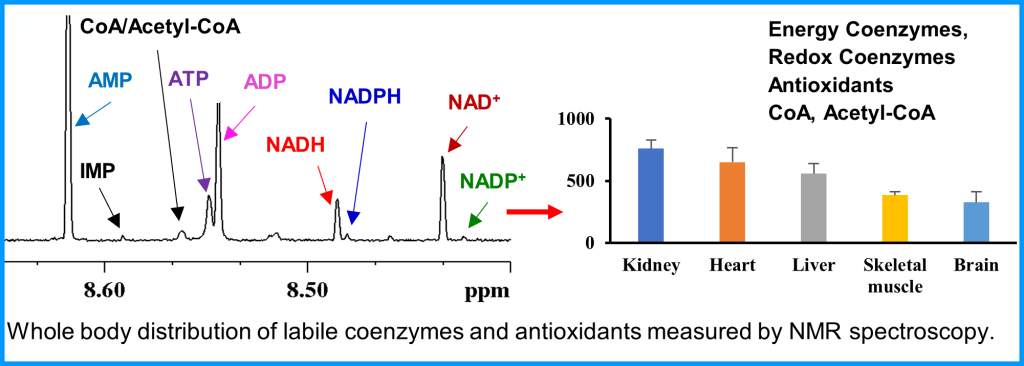
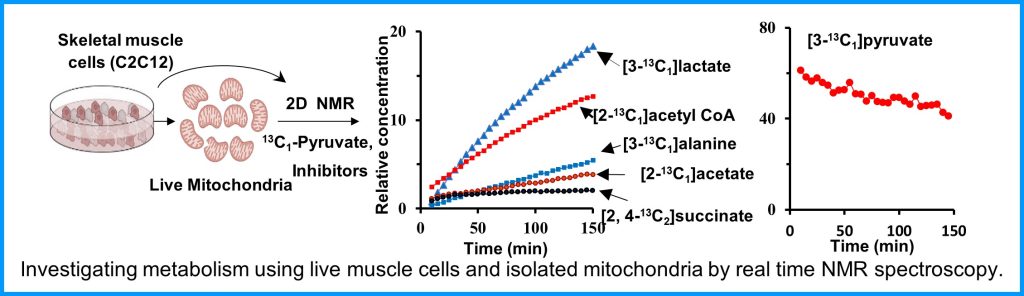
Metabolism and mitochondrial function are at the center of muscle health and degeneration. Mitochondrial dysfunction is a key component of many skeletal muscle degenerative disorders, including muscular dystrophy, disuse atrophy, and sarcopenia. New capabilities, in the form of metabolite profiling, provide advanced methods to investigate muscle metabolism. Metabolomics approaches, which focus on the quantitative analysis of large numbers of metabolites in complex bio-specimens, are providing a wealth of new data. Metabolites (<1000 Da), represent the end products of gene, transcripts and protein function, and provide an instantaneous snapshot of biological status. To date, numerous pathological aspects of muscle metabolism including myopathy disorders, endocrine disorders and consequences of aging, have been investigated using metabolomics methods.
The conditions such as mitochondrial oxidative stress, apoptosis and cellular stress responses have all been implicated in skeletal muscle atrophy and degeneration. However, despite the growing interest and research focus, the mechanistic links between skeletal muscle dysfunction and mitochondrial function remain unclear.

With a focus of bridging the knowledge gap between mechanistic understanding and skeletomuscular diseases, the Metabolism Core offers novel tools and resources for investigation of cellular and subcellular functions under normal and a wide variety of muscular disease conditions. Additionally, the Metabolism Core offers Training and Pilot Studies to encourage new users and enable current collaborators to access the state-of-the-art resources that are available. The Core develops unique scientific tools that expand access to a variety of complementary methods for measuring metabolism and in vivo mitochondrial function to the broader scientific community. Access to these tools will allow diverse groups to move beyond traditional biochemical characterization of the role of mitochondria in skeletal muscle disease and therapies. Our increased access to diverse models of skeletal muscle disease will also provide insight into common disease mechanisms by providing a comparative dataset to identify whether there are general mechanisms that lead to successful adaptation or failure of energy homeostasis and disease. The integration of in vivo measurements of mitochondrial function and energetics with ex vivo analysis of oxidative phosphorylation and metabolism via metabolomics are ideally suited for the investigation of skeletomuscular diseases.
Facilities and Resources Available
The CTMR Metabolism Core liaises closely with the Northwest Metabolomics Research Center.

Interested in utilizing our services?
Please contact Core Director, Dan Raftery (draftery@uw.edu), to discuss your project and learn more about how our services can support your project’s muscle metabolism needs.
Personnel
Dr. Daniel Raftery, Core Director, is responsible for managing the scientific operations of the Metabolism Core. Dr. Raftery directs the research activities of the metabolomics work, including the targeted and global MS and NMR spectroscopy, initial data processing, and assists with the interpretation of the metabolomics results. Dr. Raftery is also responsible for coordinating with other cores for projects involving multiple CTMR resources.
Dr. David Marcinek, Core Associate Director, is responsible for overseeing all aspects of in vivo and ex vivo mitochondrial analysis, including scheduling and prioritizing MR instrument usage, data quality control, and training of new scientists in the analysis of mitochondrial function.
Dr. Hannele Ruohola-Baker has extensive experience in mitochondrial metabolism and the use of the Seahorse analyzer that is offered as a service to Metabolism Core users. She is also very experienced with cell biology and metabolism.
Dr. Nagana Gowda is the key initial contact for users of the Core, and works with the other Core staff members to integrate the measurements and data across the different platforms and capabilities offered within the Core. Dr. Gowda has extensive experience in metabolomics and its application to a wide set of biological studies including biomarker discovery and cellular metabolism studies.
Dr. Karin Fischer has extensive experience with metabolism measurements using the Seahorse analyzer, and is responsible for all such measurements on the instrument, and assists users with sample preparation and data interpretation.
Cheryl Chan is a Research Scientist with extensive experience with metabolism measurements using the Seahorse analyzer and is responsible for all such measurements on the instrument. She will provide assistance to Core users in the proper sample preparation and will also help users interpret their data.
Dr. Matthew Campbell, Acting Assistant Professor, has 15 years of experience working in models of skeletal muscle disease and has focused on the mitochondrial and metabolic deficits associated with skeletal muscle dysfunction in aging and chronic disease working with Dr. Marcinek. He is responsible for maintaining the Oroboros Power O2K respirometer, coordinating projects involving in vivo and ex vivo mitochondrial assays, training external users and working with Dr. Marcinek to maintain quality control on all mitochondrial function data for the Metabolism Core.
Overview
The Quantitative Analysis Core provides computational modeling and statistical analysis tools and services to investigators in the CTMR. This core provides computational models at the level of single molecules (molecular dynamics) and at the level of the sarcomere. We also seek to meld multi-scale computational tools and visualization software to be used in multi-scale research on metabolism (energy supply) and contractile mechanics (energy demand/use), how these change in developing muscle and how they are altered in skeletal muscle disease. This core is also seeking to build tools for machine learning algorithms applied to both experimental data and computational simulations.
Molecular dynamics models focus on protein simulation models and computational tools for atomic-molecular level analysis of muscle proteins to gain a molecular framework for interpretation of experiments and for future experimental design.
Sarcomere scale models focus on Monte-Carlo simulations of force generation and energy utilization during muscle activation with a focus on the mechanochemistry in the myofilament lattice. Contraction and metabolic models are valuable for mechanistic interpretations of experimental data, informing experimental design and providing predictive power in translational muscle research. All relevant Python Code is now available at our Github repository: https://github.com/travistune3/multifil_five_state
Services
Interested in utilizing our services?
Please contact Program Manager, Katie Mitzelfelt (kmitz@uw.edu), to schedule a time to discuss your project and learn more about how our services can support your project’s muscle computational and statistical analysis needs.
Personnel
Dr. Jennifer Davis, Core Co-Director, is responsible for managing the scientific and financial operations of the Core. She will direct the research activities of the Core and assist in coordinating computational efforts among the subgroups of this core and with the University of Washington e-Science Program. Dr. Davis has over 20 years of experience in skeletal and cardiac muscle biology with a focus on the regulation of muscle contraction, remodeling, and regeneration. She has implemented numerous models at the sarcomeric and cellular scale to examine fundamental problems in striated muscle biology, and her research readily employs machine learning and data science techniques to identify transcriptional, morphologic, and mechanical features driving pathologic processes in striated muscle.
Dr. David Beck, Core Co-Director, works closely with Dr. Davis regarding all aspects of Core including recruiting users, overseeing personnel involved in Core D activities, and coordinating and communicating Core D activities. Dr. Beck is also the Director of Research at the UW eScience Institute and has expertise in biophysical chemistry software, methods, and application development. Through the eScience Institute, he leads the development of workflow design and engineering for data-intensive analytics, data mining, and strategies for large biological datasets that are invaluable for our faculty and Cores requiring large data management and analytics expertise and tools.
Dr. Matthew Childers is a Research Scientist with 10 years of experience in bioengineering and applied science, and training in
molecular dynamic simulations. He performs molecular dynamic and atomistic modeling for the Core, as well as contributing effort towards
integrating atomistic, subcellular, and cellular models. Dr. Childers will work with students and faculty to progress seed projects.
Dr. Sage Malingen is a postdoctoral fellow with expertise in mathematics and training in both sarcomere mechanics and modeling cell membrane mechanics. She works on integrating atomistic, subcellular, and cellular models to better understand muscle structure and
function for Core. Dr. Malingen will work with students and faculty to progress seed projects.
Dr. Travis Tune is a Research Scientist with a background in physics and data science and expertise in computational modeling of complex biological systems. He works on sarcomeric and celular models for the Core, as well as data science and machine learning approaches to tackle inverse problems in muscle biology. Dr. Tune will work with students and faculty to progress seed projects requiring his expertise
Additional Resources from CTMR Partners

A partner of the CTMR, Seattle Children’s Hospital clinical laboratory, offers Mitochondrial Respiratory Chain Enzyme Analysis in muscle.

Specific ordering information can be found online here. The Biochemical Genetics test requisition form is available online here.
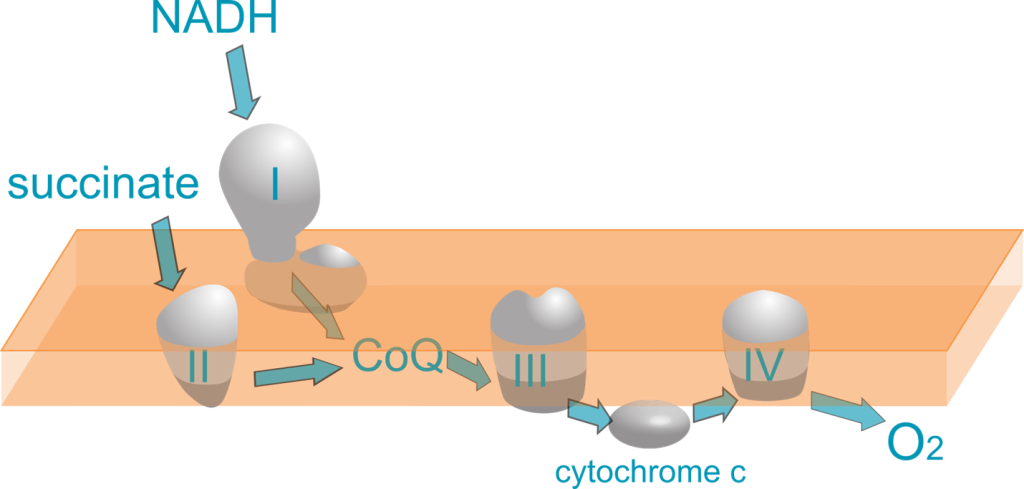
Mitochondrial Respiratory Chain Enzyme Analysis can provide supporting evidence for a diagnosis of mitochondrial disease in patients with clinical symptoms and negative or inconclusive DNA results. Mitochondrial Respiratory Chain Enzyme Analysis was validated using residual muscle tissue received by the pathology/histology laboratory at Seattle Children’s. The function of Complexes 1 through 4 are assessed using specific inhibitors and electron acceptors that change color with redox state (Agilent, Cary 6 UV-Vis Spectrophotometer).
We offer testing services for model organisms and alternative sample types to support research. Clinical validation of cultured fibroblasts is in process.
Please contact Dr. Anna Scott (Anna.Scott@seattlechildrens.org) in the Biochemical Genetics Laboratory with any questions.

